- Research
- Open access
- Published:
QSAR, pharmacophore modeling and molecular docking studies to identify structural alerts for some nitrogen heterocycles as dual inhibitor of telomerase reverse transcriptase and human telomeric G-quadruplex DNA
Future Journal of Pharmaceutical Sciences volume 7, Article number: 231 (2021)
Abstract
Background
Telomerase reverse transcriptase (TERT) and human telomeric G-quadruplex DNA are amongst the favorable target for researchers to discover novel and more effective anticancer agents. To understand and elucidate structure activity relationship and mechanism of inhibition of telomerase reverse transcriptase (TERT) and human telomeric G-quadruplex DNA, a QSAR modeling and molecular docking were conducted.
Results
Two robust QSAR model were obtained which consist of full set QSAR model (R2: 0.8174, CCCtr: 0.8995, Q2loo: 0.7881, Q2LMO: 0.7814) and divided set QSAR model (R2: 0.8217, CCCtr: 0.9021, Q2loo: 0.7886, Q2LMO: 0.7783, Q2-F1: 0.7078, Q2-F2: 0.6865, Q2-F3: 0.7346) for envisaging the inhibitory activity of telomerase reverse transcriptase (TERT) and human telomeric G-quadruplex DNA. The analysis reveals that carbon atom exactly at 3 bonds from aromatic carbon atom, nitrogen atom exactly at six bonds from planer nitrogen atom, aromatic carbon atom within 2 A0 from the center of mass of molecule and occurrence of element hydrogen within 2 A0 from donar atom are the key pharmacophoric features important for dual inhibition of TERT and human telomeric G-quadruplex DNA. To validate this analysis, pharmacophore modeling and the molecular docking is performed. Molecular docking analysis support QSAR analysis and revealed that, dual inhibition of TERT and human telomeric DNA is mainly contributed from hydrophobic and hydrogen bonding interactions.
Conclusion
The findings of molecular docking, pharmacophore modelling, and QSAR are all consistent and in strong agreement. The validated QSAR analyses can detect structural alerts, pharmacophore modelling can classify a molecule's consensus pharmacophore involving hydrophobic and acceptor regions, whereas docking analysis can reveal the mechanism of dual inhibition of telomerase reverse transcriptase (TERT) and human telomeric G-quadruplex DNA. The combination of QSAR, pharmacophore modeling and molecular docking may be useful for the future drug design of dual inhibitors to combat the devastating issue of resistance.
Graphical abstract

Background
Transcriptase has emerged as a possible drug target in cancer therapeutics because of following whys and wherefores [1]: Every human being has telomerase, which is active in the early stages of life to preserve telomere duration. This will maintain the chromosomal integrity of recurrently dividing cells, which is supposed to be inactive in most somatic cells and is maintained during adulthood [1]. The telomerase enzyme is found in 80–90% of cancer cells isolated from major human tumors, although it is not found in the adjacent cells of healthy human tissue [2, 3]. Given these evidences and particulars, telomerase has gotten a lot of attention as a potential target for developing new anticancer drugs.
Telomerase is an RNA–protein complex (RNP) that comprises 30 ends of linear chromosomes which give rise to generation of the small telomere-repeat sequence (TTGGGG in ciliates and TTAGGG in humans) by using RNA prototype. The RNA prototype is a component of its essential telomerase RNA (TER) component and its basic telomerase reverse transcriptase (TERT) [4]. Telomerase activity is strongly controlled throughout development and oncogenesis [5]. The reported evidences for telomerase makes it a popular therapeutic target, as well as inhibitory agents with potential for cancer treatment [6]. Many hTERT inhibitors were reported [2, 7], and some of them, comprising BIBR1532 [8,9,10] showed promising anticancer effects [11,12,13].
In silico development of new molecules by using a logical structure-based drug design approach that identifies prominent hits and confirms activity for carefully selected hits. As a result, finding such hits using a hybrid ligand- and structure-based drug design approach will aid in the balanced development of more effective telomerase inhibitors [14, 15].
Furthermore, noncoding repetition orders of the guanine-rich DNA, which are important for preventing the cell from recombination and degradation, are found at the telomeric ends of the chromosomes. Telomere are the extremities of eukaryotic chromosomes, are necessary for maintaining genome integrity and appear to play crucial a role in cellular aging and the cancer.
They consist of tandemly replicated DNA sequences with a G-rich strand directed 5′ to 3′ towards the chromosome's end [13]. The telomerase activation is associated with the shortening of telomere, tracked by the activation of the DNA destructive responses. This involves the cell cycle arrest, senescence and apoptosis [16,17,18,19,20,21,22]. Telomerase is an imminent anticancer drug target which is supported by the evidence that its activity is described and reported in 85–90% of all human tumors, but not in normal cells [23]. The Cell cycle arrest and apoptosis effects have rendered telomerase as a striking target in the field of anticancer drug discovery and novel therapeutics [24,25,26,27,28].
To date, a diverse array of G-quadruplex-stabilizing compounds has been investigated and reported by various researchers, including macrocyclic oxazolo [29,30,31,32,33], anthraquinones [34, 35], acridines [36,37,38,39,40], cationic porphyrins [41,42,43,44,45,46], bistriazoles [47], perylenes [48,49,50], ethidium derivatives [51, 52], fluorenones [53], pentacyclic acridinium salts [54,55,56], and fluoroquino phenoxazines [57,58,59,60,61,62].
Quantitative structure–activity relationship (QSAR) studies attempt to find new and similar molecules in the broad databases of reported molecules with known established observed activities or properties [63,64,65]. The discovery of such a statistical correlation opens up the possibility of predicting the activities and properties of new compounds and, as a result, guiding the synthesis of new molecules without having to implement it.
Multidrug resistance (MDR) is one of the major concerns associated during the course of anticancer treatment. On the other hand, the inhibition of a single target repeatedly display momentary effectiveness due to emergence of the drug resistance [66]. Perceiving that cancers are heterogeneous entities, the simultaneous inhibition of multiple targets is needed to obtain the optimal effect. As a result, finding new and safe dual inhibitors is critical to overcoming the resistance issue in cancer treatment. The main emphasis in current study is to build a QSAR model which find the various structural alerts and the features in nitrogen heterocycles containing compound and their correlation with telomerase reverse transcriptase (TERT) and human telomeric G-quadruplex DNA inhibition. Furthermore molecular docking, may perhaps to be used to understand the dual inhibitory mechanism and interactions between the ligands and receptor for predicting their binding affinity. In addition, pharmacophore modeling is used to reveal consensus pharmacophoric features required for the dual inhibition of TERT and human telomeric G-quadruplex DNA.
Method
The QSAR experimental methodology consist of selection of dataset, calculation of molecular descriptors, feature selection algorithm, validation process, and correlation in relation to structural landscapes (i.e. OECD guidelines). The main goal of using experimental methodology is to build a QSAR model with a good equilibrium of external predictive capability (quantitative/predictive QSAR) and understanding of QSAR model as well as molecular descriptors in terms of structural alerts responsible for biological activity (qualitative/descriptive QSAR) [67,68,69,70,71,72,73].
Thus, classic method has been followed to develop a stable QSAR model for inhibitory activity of nitrogen ring containing heterocycles for TERT and human telomeric G quadruplex DNA. More particulars about the technique followed in the current work are available in the literature [71, 74,75,76,77,78].
Selection of dataset
In this study, a dataset of structurally varied 82 nitrogen ring containing heterocycles experimentally tested against Human TERT inhibitory potential has been carefully chosen for QSAR investigation from renown and publicly accessible ChEMBL database (https://www.ebi.ac.uk/chembl/). The dataset includes a wide range of molecules with different substituents, such as acridine rings, triazoles rings, pyrimidine rings, and so on. As a result, in order to create a robust QSAR model, we try to cover as much chemical space as possible. The experimentally reported EC50 values range from 2 to 23,500 nM, which were transformed to pEC50 (− log10EC50) prior to QSAR model building. The SMILES notations, EC50 and pEC50 for selected nitrogen heterocycles as an example only are presented in Table 1. In Table 1, we have depicted five most and least active compounds as representative examples only. In addition, the common scaffolds have been presented in Fig. 1 (Presentation of Serial number, CheMBL id, Smiles and Pec50 value of 82 compounds is given in Additional file 1: Table S5).
The 2D-structures of all 82 compounds were sketch by ACD ChemSketch Freeware (www.acdlabs.com) tracked by conversion to 3D structures using Avogadro ver. 1.02 (https://avogadro.cc/) by means of MMFF94 force field for geometry optimization and partial charge assignment. The resulting parameters were used for geometry optimization: Force Field: MMFF94, Algorithm: Steepest Descent, numeral of steps used for optimization: 1000.
Calculation and pruning of molecular descriptors
PyDescriptor were employed for molecular descriptor calculation using 3D-optimized structures of dataset compounds. PyDescriptor are available as plugin in PyMOL molecular viewer which calculated a bunch of more than 29,000 molecular descriptors for each molecule used in present QSAR analysis [71]. In the next step, we have implemented objective feature selection (OFS) to minimize the group of molecular descriptors. The OFS method intricate removal of persistent, nearly persistent (95% molecules) and greatly correlated molecular descriptors (R > 0.90). OFS lead to condensed bunch of 494 molecular descriptors, which still encompasses comprehensive range of PyDescriptor because of incidence of 1D- to 3D-molecular descriptors. This reduced descriptor pool was further used for the subjective feature selection (SFS) [71, 78,79,80] (Calculated Py-Descriptor values used to build QSAR model is depicted in Additional file 1: Table S6).
Subjective feature selection (model building)
The method of SFS was executed by using QSARINS-2.2.4 with default setting in which a number of generations was set to 1000. The genetic algorithm (GA) module available in QSARINS-2.2.4 employ Q2 as a fitting parameter to circumvent over fitting and insertion of redundant variables during model building. A key decision in evolving a successful QSAR model is to stop adding molecular descriptors to the model at appropriate point of time. In the current study, breaking point is obtained by means of graph was drawn amid the number of descriptors intricated in the models and Q2 value. In the conclusion, the number of molecular descriptors conforming to the breaking point was considered optimal for model development. The graph amid numbers of variables used in the models against Q2 value is shown in Fig. 2. From Fig. 2, it is clear that the breaking point links to five variables. Therefore, five descriptor were used to derive robust QSAR model while QSAR models with more than five descriptors were excluded [79, 81].
To gain deep understanding of structural features prominent for Dual inhibition of telomerase reverse transcriptase as well as human telomeric G DNA, we are expected to develop (Descriptive QSAR) statistically satisfactory GA-MLR originated QSAR model [82, 83].
To do so, we used the strategy of dividing the entire dataset into two sets: 80% training set and 20% prediction set. Using this technique, training set give rise to the most relevant and appropriate number of molecular descriptors for QSAR model, while the prediction set (test set) was employed for external validation, i.e., to ascertain the external prediction capability (predictive QSAR). The plan has been portrayed in Fig. 3.
Validation of QSAR model
Following the development of a QSAR model, it is extremely crucial and important to validate the model for external predictive potential in order to determine its performance and scope for predicting biological activity in lead/drug optimization during the drug discovery phase [56, 57, 73]. As a result, not only were extensive internal validations and the Y-scrambling technique used, but an external prediction range of 20% molecules was also used to verify the model's statistical robustness. A Williams plot was also created to ascertain the applicability domain of the developed QSAR model. Additionally, the guidelines were used to choose and validate a QSAR model. R2tr 0.6, Q2loo 0.5, Q2LMO 0.6, R2 > Q2, R2ex 0.6 RMSEtr < RMSEcv, DK 0.05, CCC 0.80, Q2-Fn 0.60, r2m 0.5, (1 − r2/ro2) < 0.1 0.9 k 1.1 or (1 − r2/r′o2) < 0.1, 0.9 k′ 1.1, ro2 r′o2 < 0.3 with RMSE and MAE as low as possible. The Q2LMO value stated here is mean value of 2000 repetitions with 30% of the population (molecules) arbitrarily excluded from the training set at each time. The external predictive capacity of model was find out by using external validation parameters, viz., RMSEex MAEex, R2ex, Q2F1, Q2F2, Q2F3, and CCCex. All QSAR models that do not achieve the endorsed lower-limit values for above statistical parameters have been directly excluded,
The inter-correlation between descriptors were tested by the QUIK rule (Q under Influence of K). QUICK rule was fixed to 0.05 to lessen inter-correlation among descriptors. The reliability of the developed QSAR model was ascertained by Y-randomization set at 2000 iterations to check the fitting of the randomly reordered Y-data. For the randomization of the build QSAR model, the dependent variables (PIC50 value) of the training set have been shuffled and new coefficients of determination were calculated. The significantly low value of the coefficients of determination of the new models specify that the reported model in the present QSAR analysis is not obtained by chancy correlation. The external validation of all the models were verified with the subsequent validation criteria: r2ext (external determination coefficient), Q2F1, Q2F2, Q2F3, concordance correlation coefficient (CCC), CCCext, r2 m, and Δr2 m. The parameter R2 m (overall) penalizes a model for large differences between observed and predicted values of the compounds of the whole set (considering both training and test sets). The Δr2m estimated the indulgent between the values of the predicted and the resultant experimental activity data (PIC50 value). It has been reported that, the observed value for the Δr2m should be preferentially lower than 0.2 provided that, value of r2m > 0.5.
Additionally, all the QSAR models were evaluated for validation parameters, such as Golbraikh and Tropsha’s criteria to justify model reliability and robustness. Generally, good predictive ability of the developed QSAR model depends upon closeness of predicted value against observed (experimental biological activity) value. Even, presence of single outlier diminish the predictive capability of the developed QSAR model. Subsequently, we have tried to highlight the outlier on the basis of those compounds who showed significantly high residual value in GA-MLR QSAR models. Moreover, we have identified the outlier compounds by comparing the predicted value with three standardized residual values. Likewise, structural variation in database compounds was observed by leverage effect in Williams plot. The applicability domain of the developed QSAR model is ascertained by merging the leverage and the standard residuals [68, 69, 72, 74, 84, 85].
Pharmacophore modeling
To achieve a consensus pharmacophore model, we have generated lowest energy conformer of the most active compound 82 that was used to align all of the molecules in the dataset. Then, LIQUID 1.0, a free PyMOL plugin, was used to produce consensus pharmacophore model expending default settings. Whereas, in Fig. 12, the pdb files (5cqg) were obtained from publicly accessible databank (www.rcsb.org). Afterward, the bound pdb ligands were isolated without any alteration or optimization, that is, the X-ray crystallographic resolved crystal structure of extracted ligand was used as it is to generate the pharmacophore model with LIQUID 1.0 [86] (Fig. 4).
Docking of the inhibitors
Molecular docking studies were performed using the NRGSuite software package. The NRGSuite package equipped with FlexAID that contains four primary panels to specify the input target protein and ligand to be docked, configuration of the target and ligand and simulation. All the crystallized water molecules and coordinated molecules were found in the crystal structure of BIBR1532 anchored with the Tribolium castaneum catalytic subunit of the RNA prototype the telomerase (tcTERT) were preserved (pdb id-5cqg) during a docking procedure. The binding site residues of tcTERT was ascertain by using site finder function in NRGSuite to determine active site in crystallized ligand BIBR1532 with the extraction of ligand structure present in pdb for redocking purpose. Site finder option reveals two binding site in chain A and B where crystallized ligand BIBR1532was found to be bound. As a result, the inhibitor compound 82 and the co-crystallized ligand were docked using NRGSuite’s site finder function. The FlexAID uses genetic algorithm. A number of important parameters, notably the number of chromosomes and generations can be defined in this panel. Additionally, the number of top results that are visualized during the simulation and the frequency (in numbers of generations) to refresh the visualization can be set. Then, the poses obtained during the placement stage were then fine-tuned using the Induced Fit method, which allows for protein versatility during ligand binding and thus improves the interaction prediction accuracy. The top five uppermost scoring poses were then achieve with the GBVI/WSA dG scoring utility. The final performance was analyzed, and docked poses inside the binding site that were not properly oriented (for catalytic site) and whose versatile alignments were compared to the top scoring pose of compound 82 were included. The docking poses of compound 82 were then superimposed on top of the co-crystallized ligand BIBR1532 to determine the most favorable docked conformation for telomerase inhibition.
Results
Full set model
PEC50 = 4.191 (± 0.596) + 0.407 (± 0.09) * fdonH2A + − 0.251 (± 0.095) * ringN_acc_8A + 0.261 (± 0.131) * faroCC3B + 0.214 (± 0.099) * fplaNN6B + 0.263 (± 0.065) * com_aroC_2A++
R2: 0.8174, R2adj: 0.8054, R2–R2adj: 0.0120, LOF: 0.1499, Kxx: 0.2604, Delta K: 0.1186, RMSEtr: 0.3400, MAEtr: 0.2510, RSStr: 9.4780, CCCtr: 0.8995, s: 0.3531, F: 68.0476, Q2loo: 0.7881, R2–Q2loo: 0.0293, RMSEcv: 0.3663, MAEcv: 0.2702, PRESScv: 11.0009, CCCcv: 0.8831, Q2LMO: 0.7814, R2Yscr: 0.0597, Q2Yscr: − 0.0987, RMSE AV Yscr: 0.7714. (Depiction of QSAR results along with their experimental and predicted EC50 values for full set model is given in Additional file 1: Table S4).
Divided set model (80:20)
PEC50 = 5.355 (± 0.563) + 0.367 (± 0.1) * fdonH2A + − 0.287 (± 0.098) * ringN_acc_8A + − 0.162 (± 0.119) * notringC_aroC_2B + 0.336 (± 0.127) * fplaNN6B + 0.274 (± 0.071) * com_aroC_2A++
R2: 0.8217, R2adj: 0.8068, R2–R2adj: 0.0149, LOF: 0.1600, Kxx: 0.2783, Delta K: 0.0936, RMSEtr: 0.3394, MAEtr: 0.2636, RSStr: 7.6042, CCCtr: 0.9021, s: 0.3560, F: 55.2934, Q2loo: 0.7886, R2–Q2loo: 0.0331, RMSEcv: 0.3696, MAEcv: 0.2882, PRESScv: 9.0164, CCCcv: 0.8842, Q2LMO: 0.7783, R2Yscr: 0.0777, Q2Yscr: − 0.1253, RMSE AV Yscr: 0.7717, MSEext: 0.414, MAEext: 0.3415, PRESSext: 2.7438, R2ext: 0.7020, Q2-F1: 0.7078, Q2-F2: 0.6865, Q2-F3: 0.7346, CCCext: 0.8354, r2m aver.: 0.5878, r2m delta: 0.0810.
In the present QSAR analysis, compound 82 was detected as X outlier while compound 8, 13 were depicted as high leverage influential (see Fig. 5). Moreover, apart from fitness function Q2, we have displayed another fitness function of concordance correlation coefficient (CCCext) as one of the external validation parameter. (Depiction of QSAR results for divided set model along with their experimental and predicted EC50 values for divided set model is given in Additional file 1: Table S3).
Discussion
faroCC3B
The descriptor faroCC3B point out frequency of occurrence of carbon atom exactly at 3 bonds from aromatic carbon atom. Since this descriptor has positive coefficient, this means that, increase in the value of this descriptor will increase its pEC50 value for the molecules used in present study. In compound 81, C3 and C6 propanamide substituent placed at a topological distance of 3 bonds from C3 and C6 aromatic carbon atoms of acridine ring, amino methyl carbon atom placed at C2 postion of pyrrolidine ring is separated by the topological distance of three bonds from C9 aromatic carbon of acridine ring and C3, C4 carbon atoms of pyrrolidine ring is placed at a topological distance of three bonds from C9 aromatic carbon of acridine Ring. Same pattern of descriptor faroCC3B is observed in compound 7 except pyrrolidine ring is replaced by amino cyclopropyl substituent on C9 position of acridine ring. If we compare activity profile of compound 81 and 75, we observed that pyrrolidine substituent is more favorable for anticancer activity rather than amino cyclopropyl substituent. This observation highlight the variation in the activity of compound 81 and 75 in nanomolar range.
To add more, both aliphatic chain and unsaturated centers in molecule significantly contributed to the overall lipophilicity of the molecule. Therefore, lipophilicity is the key feature that govern dual inhibition of TERT and human G DNA. Subsequently, attachment of pyrrolidine ring to the acridine ring through the single bond impart enough flexibility to the compound 81 therefore, sterically lock it into the active conformation within TERT and human telomerase G DNA.
Ranganathan et al. and Khanna et al. have reported that, most of the molecules in the metabolite dataset used studies contains a carbon atoms in the range 35–55 which is 32% i.e. 5–25 carbon atoms per molecule. The carbon atom content in metabolites has a mean of 33 atoms and maximum up to the 100. On the other hand, drugs molecule have an average of 18 carbon atoms per molecule, with a maximum of 256 and 76% of drugs consist of carbon atoms in the range of 5–25 [85]. In QSAR model descriptor faroCC3B highlight the importance of occurrence of carbon atom exactly at 3 bonds from aromatic carbon atom in dataset molecules (see Fig. 6).
Further, in compound 66, descriptor faroCC3B is observe at C3and C6 aromatic carbon of acridine ring which reveals that, decrease in the cloud of carbon atoms placed at topological distance of three bonds from the aromatic carbon atom, further diminishes the anticancer activity of compound 66. For increasing anticancer profile of compound 66, substitution of pyrrolidine ring with an amino methyl substitution at C9 position of aromatic carbon of acridine ring is recommended. This observation supported the fact that, compound 81and 75 have five such carbon atoms placed at a topological distance of three bonds from aromatic carbon atoms while compound 66, 45 and 8 shows two centers except compound 13 in which carbon with topological distance of three bond from aromatic carbon atom is missing, therefore it is clear that variation in pEC50 value is due to absence of carbon atoms at a topological distance of three bonds from aromatic carbon atoms. Here greater the number of carbon atom at a topological distance of three bonds from aromatic carbon atom, higher will be the anticancer activity of stated compounds under study. Enhance cloud of carbon atom augments lipophilicity which in turn indicate maximum hydrophobic interaction with receptor.
fplaNN6B
The descriptor fplaNN6B stand for the frequency of occurrence of nitrogen exactly at six bonds from planer nitrogen atom. The positive coefficient designates that an increase in the number of such Nitrogen atoms may plausibly enhances the anticancer activity (pEC50 value). Ranganathan et al. and Khanna et al. have recognized that drugs molecules clearly possess the maximum number of the nitrogen atoms, followed by toxin molecules and lastly, metabolites [85].
Pennington et al. established the importance of nitrogen in heterocyclic compounds. Pennington et al. reported that the replacement of a CH group with N atom in aromatic and hetero aromatic ring structures can have many beneficial effects on molecular and physicochemical properties and intra and intermolecular interactions that may give rise to improved pharmacological profiles in drug discovery. Moreover, Pennington et al. also investigated that, a N atom in aromatic and heteroaromatic ring systems can influence the number of intra- and intermolecular orbital, steric, electrostatic, and hydrophobic interactions such as lone pair, dipole–dipole, hydrogen bonding, metal coordination, van der Waals, σ-hole, σ*S − X, and π-system interactions, which in turn can translate to modified pharmacological profiles [87].
This observation is reinforced by a simple comparison of the subsequent pair of molecules: comp-81 (PEC50-7.74 nm) with comp-40 (PEC50-6.59 nm), comp-80 (PEC50-7.74 nm) with comp-45 (PEC50- 6.67 nm) (see Fig. 2). In case of compound 2, this feature is missing. Therefore, we can say that presence of planer nitrogen is most important for augmenting biological activity performance of molecule (see Fig. 7). In our dataset, compound 62 to 82 contains two planer nitrogens, compound 40, 45, 46, 52, 67, 78, 82 consist of one planer nitrogen while planer nitrogen in absent compounds 1 to 4 and 6 to 32. Here, nitrogen becomes planar when its lone pair becomes involved in pi-bonding. The five-membered rings have significant delocalization of electrons to produce a cloud system similar to that in benzene. As a result, planer nitrogen increases the electron cloud in the molecule, which strengthens electrostatic interactions with the receptor surface by exhibiting pi bonding.
ringN_acc_8A
This descriptor depicts occurrence of ring nitrogen within 8 A0 from the acceptor atom. In QSAR model, this descriptor has negative correlation with PEC50 value. Therefore, number of ring nitrogen within 8 A0 from acceptor atom must be retained, as low as possible to enhance the anticancer activity (PEC50 value). Increase in the value of descriptor ringN_acc_8A will further decreases the anticancer activity profile of the compounds in dataset. This is observed when PEC50 of compound 23 compared with compound 41. This could be the possible reason for the difference in the PEC50 value of compound 23 and 41 (see Fig. 8).
In general, it is established that, close combination of ring nitrogen and acceptor is avoided to prevent intramolecular hydrogen bonding in the molecule. Specially, when oxygen and nitrogen are connected by single bond to the neighbor atoms. Thus, the descriptor ringN_acc_8A provides a hint to avoid close proximity of ring nitrogen with acceptor atom to avoid the prospect of intramolecular bonding.
com_aroC_2A
The descriptor com_aroC_2A specifies occurrence of aromatic carbon atom within 2 A0 from center of mass of molecule. This descriptor is positively correlated with PEC50. Hence, this value must be kept as high as possible. In case of compound 82 (PEC50 = 8.69 nm), there are around six carbons present within the radius of 2 A0 from the center of mass of the molecule while in compound 19, only three carbons are present within 2 A0 from center of mass of molecule. Therefore, it is reasonable to settle that, difference in the activity of stated compound is due to the number of carbons present within 2 A0 from the center of mass (see Fig. 9).
The same is true for compound 13, which has five carbons within 2 A0 of the center of mass. It is reasonable to conclude that the activity of compounds 82 and 13 differs solely due to the number of carbons within 2 A0 of the center of mass, and that this may be the cause of the differences in the activity profiles of both molecules.
This could be the possible reason for the differences in the activity profile of both molecules. In general, the presence of aromatic carbon atom affect overall lipophilicity of the molecule, therefore, it is rational to predict that, carbon atoms present in the vicinity of center of mass plays crucial role in hydrophobic interactions with the receptor.
notringC_aroC_2B
The descriptor notringC_aroC_2B describes the occurrence of non-ring carbon atom exactly at or within 2 bonds from aromatic carbon atoms, providing different level and type of useful information. Since this descriptor has a negative coefficient in the model, raising its value can result in a lower activity profile. In this case, a compound with a higher number of non-ring carbon atoms exactly at or within two bonds from aromatic carbon atoms might have lower activity than one with fewer of these aromatic carbons. This is observed when comparing compound 37 to 61 and compound 33 to 78 (see Fig. 10). This finding supports the fact of the variation in activity of the stated compounds.
In compound 78, non ring carbon atom containing amide group is present at terminal position, therefore, it is may establish that, these substituents occupy lipophilic pocket of the TERT as well human telomeric G DNA. To add more, these substituent varying steric bulk in the receptor pocket, thereby blocking the enzyme. Besides, the presence of aliphatic chain along with unsaturated pyrrolidine imparts good lipophilicity as well as flexibility to the molecule.
fdonH2A
The descriptor fdonH2A indicatesfrequency of occurance of element hydrogen within 2A from donar atom. It has positive coefficient in the developed models, therefore the number of Hydrogen atoms in the neighborhood of ring Nitrogen atoms is favorable blend to be used for lead/drug optimization. Since Hydrogen is the smallest element, it suggests that there should be minimum bulk in the vicinity of donar atoms. Therefore, in future structural modifications, steric bulk nearer to donar atoms should be circumvented to have better anticancer activity (see Fig. 11).
As the descriptor fdonH2A specifies necessity of higher number of donar feature with presence of hydrogen atom within 2 A0. In case of compound 68, five donar features are present with five hydrogen atoms within 2 A0 while four donar are reported in compound 38. As a result, it is reasonable to conclude that a greater number of donars containing hydrogen atoms is needed for greater telomerase inhibition. This may be a plausible explanation for the differences in PEC50 values among the compounds mentioned. When we compared the PEC50 values of compound 68 with 38 and compound 26 with 4, we came to the same conclusion.. In case of compound 68, steric bulk increase from amide oxygen due to butyl and hexyl aliphatic side chain. These substituents in turn augment the lipophilicity of the compound along with the selectivity towards receptor.
To add further, in compound 68, donar is capable of getting more surface area for hydrogen bonding within receptor pocket due to aliphatic side chain as compared to bulky aromatic substituent. To add more, the interaction between compound 68 and the receptor is possible due to the flexibility of aliphatic butyl side chain.
In our QSAR analysis, diverse Py molecular descriptors demonstrating dissimilar structural landscapes have provided expressive visions into the whys and wherefores for differences in the anticancer activity of dataset compounds.
Pharmacophore modeling
It is a deep-rooted and successful branch of Computer assisted drug design which is executed to recognize key structural alerts (properties) accountable for binding affinity and overall pharmacological activity of ligand. The consensus pharmacophore model displays two larger hydrophobic regions separated at a distance of 7.2 A0 and 4.2 A0 from hydrogen bond donar and two hydrogen bond acceptors placed at a distance of 2.8 A0 and 2.3 A0 from hydrogen bond donar. The pharmacophore modeling emphasized the significance of the hydrophobic nature of central acridine ring and its nearby substituents atoms. The similar observation is also reinforced by the occurrence of the descriptor faroCC3B, notringC_aroC_2B and com_aroC_2A in the QSAR model as well as recent crystal structures for BIBR1532 with TERT (see Fig. 12).
Based on a comparison of Pharmacophore model with co-crysallized ligand (pdb-5cqg) with pharmacophore model for Most active compound 82, the consensus pharmacophore model and the pharmacophore model obtained using the X-ray resolved crystal structure of extracted ligands are very close especially with respect to the presence of two large hydrophobic region (green colored) at the both end and one H-bond acceptors in the vicinity of acridine nitrogen (red colored). Thus, QSAR and pharmacophore modeling led to recognition of consensus and matching structural topographies and justified by recent crystal structure of TERT with BIBR1532. Moreover, compound 82 display hydrogen bonding and hydrophobic interaction with human G DNA, therefore, presence of hydrophobic as well as acceptor feature is crucial for binding as well as inhibition of human G DNA. The similar remark is highlighted by the occurrence of the descriptor faroCC3B, notringC_aroC_2B and com_aroC_2A in the QSAR model.
Molecular docking of compound 82 with TERT
Telomerase enzyme is a ribonucleoprotein (RNP) reverse transcriptase responsible for replicating the ends of chromosomes and sustaining genome authenticity. The TERT structure comprises four separate areas (TRBD, fingers, palm, and thumb) well-arranged into a ring thus, producing large interior binding pocket for RNA prototype and telomeric DNA during the whole process of telomere elongation. At the present, BIBR1532 molecule is in clinical trial and chemically, it is a (2-[(E)-3-naphtalen-2-yl-but-2-enoylamino]-benzoic acid). It is a non-nucleosidic, non-competitive, small-molecule inhibitor of telomerase that is regularly and constantly introduced in studies of telomerase function. (Docking results for Compound 82, BABR1532 and Epirubicin into the TERT is given in Additional file 1: Table S1).
Experimental and simulated annealing study reveals the presence of superficial but well-defined hydrophobic pocket located on the external surface of the thumb area of telomerase and following the TRBD-thumb border. This pocket is formed as a minor gap, around 10 A0 wide-ranging and 8 A0 deep, prepared by the collection of the tips and connecting loops of the helices 20, 21, and 22, 23. This cavity form as pocket and, referred as the FVYL motif/pocket, present on the well-preserved hydrophobic residues: F478, V491, Y551, and L554. The FVYL amino acid residues exert extensive hydrophobic interactions to stabilize the placement of the rings and helices near the pocket. Many well-maintained and typically hydrophobic amino acid residues occupying the interior of this pocket, which comprise M482, M483, F494, I497, W498, I550, Y551, and L554.
Examination of the BIBR1532 telomerase amalgamated crystal structure data revealed hydrogen bonding interactions amid Asn 421, Arg 433, Lys 437 of telomerase and the carbonyl and carboxylic acid groups of BIBR1532. From the docking analysis of pose 1 of compound 82display docking score of − 9.125 kcal/mol and occupied FVYL motif/pocket through pi-alkyl hydrophobic interaction with residue L554 via phenyl ring of 4-amino-phenyl substituent. Acridine nitrogen form key hydrogen bonding interaction with water molecule HOH: B735 (2.17 A0) while pyrrolidine ring carbon exhibit two carbon hydrogen bonding with residue GLY: B283 (2.56 A0, 2.81 A0) (see Fig. 13).
Meanwhile, B:MET 482 residue form pi–sulphur interactions with acridine ring, PHE: 494 execute amide–pi interaction with phenyl ring, ARG486 and ILE550 anchored alkyl hydrophobic interactions with pyrrolidine ring. In addition to this ILE497, Leu554 and ILE550 which form shape of the interior of FVYL motif/pocket and exhibit pi-alkyl hydrophobic contact with amino phenyl substituent. Compound 82 acquired same binding conformations as that of crystallized ligand BIBR1532 (see Fig. 13). As previously mentioned, we chose two binding sites 23 and 47 in the site finder choice in MOE to investigate the binding mode of compound 82 due to crystallized ligand BIBR1532 binding at two separate sites during the docking process. Docking analysis divulges that compound 82 acquired two best docked conformation with the docking score of − 9.125 kcal/mol and − 9.004 kcal/mol at first site while third docking conformation was acquired in another binding site and exhibit two hydrogen bonding interaction in which water molecule HOH: 784 (3.40 A0) bind with acridine ring and HOH: 799 (2.58 A0) attached with pyrrolidine nitrogen (see Fig. 14).
Likewise, middle acridine form electrostatic contact with ARG 486 residue through pi–cation interactions and PHE494 involved in three pi–pi stacked hydrophobic interactions with all the three acridine rings. Therefore, it is reasonable to say that acridine ring plays crucial role in enhancing binding affinity against TERT and actively involved in drug receptor interactions. In addition to this, MET482, ILE497, LEU554, ILE550, MET483, ARG486 residue from the interior lining of the FVYL motif/pocket are involved in pi-alkyl hydrophobic interactions with amino phenyl substituent and acridine ring.
Again it is not hard to see that compound 82 acquired different conformations within FVYL motif/pocket and key interaction involves hydrogen bonding and hydrophobic interactions with the involvement of water molecule and hydrophobic residues MET482, ILE497, LEU554, ILE550, MET483, and ARG486. When the docking findings were compared to those of the crystallized ligand BIBR1532, the interaction between compound 82 and amino acid residues was found to be close to that of BIBR1532 (see Fig. 15).
The descriptor com_aroC_2A andfaroCC3B pint out towards importance of lipophilicity in telomerase inhibition. Here Ligand lipophilicity influences target affinity momentarily as most discovered binding sites shows presence of at least one hydrophobic pocket in a nearby aqueous environs. The hydrophobicity give rise to the interaction between the ligands and the protein binding sites through altering the interactions between the protein and solvating waters, therefore exhibiting more promising hydrophobic interactions for both ligand and protein. Therefore, these descriptors give key information about lipophilicity is the most important factor required for telomerase inhibition and plays crucial role in monitoring the balance of hydrophobic features of molecule. Thus, it is sensible to say that docking outcomes are in complete agreement with descriptor com_aroC_2A and faroCC3B. The Descriptor ringN_acc_8A and fplaNN6B highlight the importance of ring nitrogen and planer nitrogens in QSAR model. Docking results depicted that acridine ring nitrogen (ringN_acc_8A) form pi-alkyl hydrophobic interactions with TERT receptor therefore, it is rationale to say that docking results are entirely correlated with QSAR findings. Moreover pyrrolidine ring nitrogen exhibit hydrogen bonding with water molecule which again put forth that, nitrogen atom is essential for TERT inhibition.
Molecular docking study on human telomeric G-quadruplex DNA
The human and mammalian telomeric DNAs comprises 5′-TTAGGG-3′ repeating sequences that contain numerous base pairs. Binding of small ligand to human telomeric DNA is documented to be stabilize G4 DNA, impede in functioning of gene expression/regulation is one of the strategy to develop new anticancer agents.
The established X-ray crystal structure of a human quadruplex G DNA made from four uninterrupted human telomeric DNA which repeats and developed at a K1 concentration that come close to its intracellular concentration. K1 ions were reported in the crystal structure. The folding and occurrence of the DNA in reported (pdb id-1kf1) intramolecular quadruplex, is primarily different from the Reported Na1-containing quadruplex arrangement [88, 89]. All four DNA strands are present in analogous fashion and, shows three linking trinucleotide coils placed on the outer core of the quadruplex and look like as propeller-like arrangement.
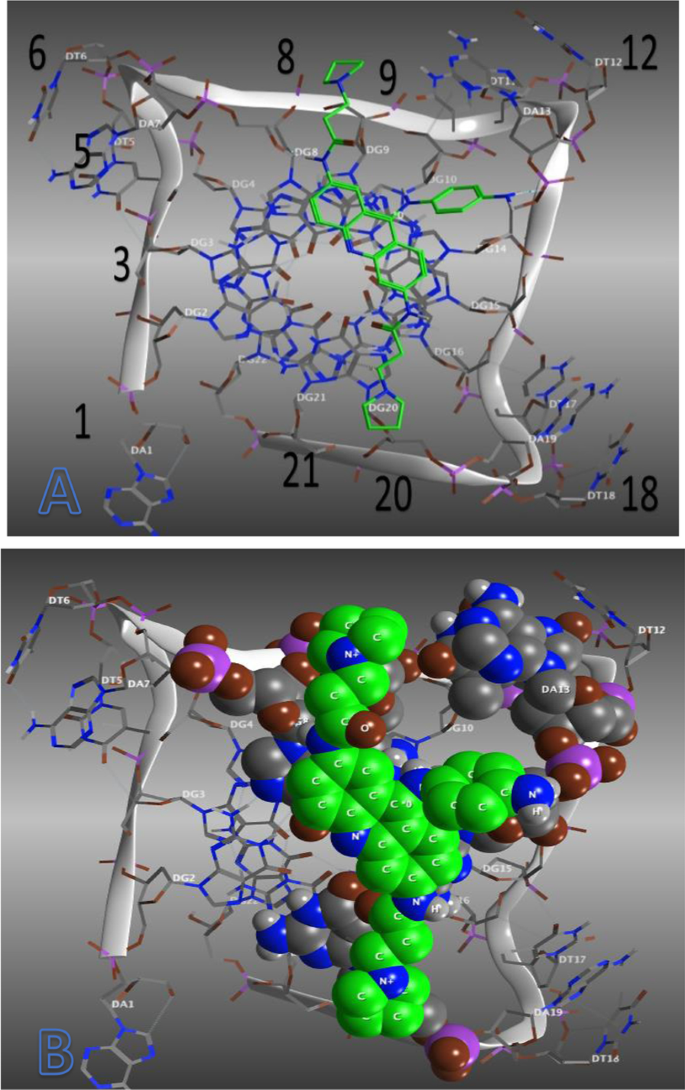
Docking studies of compound 82 in complex with human telomeric G-quadruplex DNA shows (dock score = − 7.2503 kcal/mol) that planer acridine ring loaded on the G terminal and align in between DG: 8 and DG: 9 where it exhibit pi–pi stacked interaction with DG: 8 (see Fig. 16). Here central cationic acridine ring nitrogen atom covering the central polarized carbonyl channel of negative electrostatic potential that runs through the stack of G quartets and exhibited a contact with potassium ion (K, A: 46) at topological distance of 3.34 A0. At this point, one propanamide substituent at 6 position of acridine ring orient in between DG: 8 and DG: 9 where it exhibit one hydrogen bonding interactions with water molecule HOH: 1050 (2.75 A0), one carbon hydrogen bond with DG: 8 (2.85 A0) while another propanamide substituent at 2 position stacked on DG: 20 where it form a contact of hydrogen bond with DG: 14 (2.76 A0) and carbon hydrogen bonding with DG: 20 (2.57 A0).
The central acridine ring is stabilized by pi–pi stacked interactions with DG: 8 (5.99 A0) and DG: 14 (5.61 A0). Moreover acridine ring exhibit one more pi–pi stacked contact with DG: 14 (5.13 A0) to concrete stabilization of acridine ring and human telomeric G-quadruplex complex (see Fig. 16). The binding site itself is extremely disturbed as it is appears exterior to the load of three G quartets which is connected to the channels generated from the phosphodiester backbones. (Docking results for Epirubicin and compound 82 in the human telomeric G DNA is given in Additional file 1: Table S2).
Further amide carbonyl oxygen of propanamide substituent exhibit metal acceptor contact with potassium ion (K, A: 46) at interatomic distance of 3.21 A0 which disclose close contact of 2-propanamide substituent than acridine nitrogen. Third sub-stunt 4-aminophenyl at 9th position of acridine ring align in between DG: 14 and DG: 10 where phenyl ring form pi-alkyl contact with DG: 14 (5.39 A0) while amino substituent exhibit hydrogen bonding again with DG: 14 (2.09 A0). Here it is important to note that amino substituent align very closely near DG: 12 DNA base (see Fig. 17).
Furthermore, we docked Epirubicin against human telomeric G-quadruplex and analyzed its binding orientation and modes of interaction to compare the docking findings of compound 82. The docking analysis reveals binding of Epirubicin with human telomeric G-quadruplex DNA which yielded negative docking score of − 6.1933 kcal/mol. Binding at DA: 13, DG: 14, 55, Potassium (K: 26) and 11 site is stabilized by polar and hydrophobic contacts with Epirubicin. Methoxy group substituted on 1 position align in between DG: 15 and DG: 14 where ring A, B and C form pi–pi hydrophobic contact with DG: 14 successively placed at interatomic distance of 3.70, 3.78 and 6.05 A0 (see Fig. 18). In addition to this, ring A exhibit carbon hydrogen bond with 11, 5-dione moiety form metal-acceptor contact with potassium (K, A: 26) at 3.47 A0 and 6-hydroxy substituent exert hydrogen bonding interaction with DG: 14 (2.77 A0). Moreover, ethereal linkage oxygen at 10th position exhibit covalent bond with 55, 5-hydroxy substituent form hydrogen bonding contact with 55 (2.77 A0) while 4-amino group display hydrogen bonding contact with DA: 13 at 2.20 A0.
There is distinct difference in binding of compound 82 and Epirubicin. The compound 82 was directed on the G terminal and orient in between DG: 8 and DG: 9 where it exhibit pi–pi stacked interaction with DG: 8 whereas another pyrrolidine end orient on the DG: 20 and slightly inclined near DG: 21.
Moreover, central cationic acridine ring nitrogen exhibited a metal-acceptor contact with potassium ion. In comparison to this, methoxy terminal of Epirubicin align in between DG: 15 and DG: 14 where ring A, B and C form pi–pi hydrophobic contact with DG: 14 with successive placement at interatomic distance of 3.70, 3.78 and 6.05 A0 while orientation of pyran ring of Epirubicin spread over DG: 4, DG: 8 and DG: 9 (see Fig. 19).
Conclusions
In our study, telomerase reverse transcriptase (TERT) and human telomeric G4 DNA were used as therapeutic targets to build a QSAR model. We used an 82-compound dataset that includes a variety of structures such as the acridine ring, triazoles ring, and pyrimidine ring. QSARINS 2.2 software is used to create a QSAR model using PyDescriptor and the GA-MLR method. The derived QSAR model showed high external and internal predictive ability. Compound 82 attained diverse conformations inside FVYL motif/pocket of TERT and key interaction involves hydrogen bonding and hydrophobic interactions with the involvement of water molecule and hydrophobic residues MET482, ILE497, LEU554, ILE550, MET483, and ARG486. With the bottomless analysis of docking outcomes compared with crystallized ligand BIBR1532, the interaction amid compound 82 and amino acid residues were similar to those of BIBR1532.
Moreover, molecular docking revealed that compound 82 and Epirubicin bind to G4 DNA's external grooves and loop, forming pi–pi stacking hydrophobic and hydrogen bonding associations with G DNA bases. In binding interaction of both ligands, DG: 14 and potassium ion are the common DNA base and ion, with which both ligand exhibit pi–pi stacking hydrophobic and metal acceptor interactions. In terms of orientation, both ligands terminals align by their own way on different grooves and loops which may leads to specificity of interactions with G4 DNA that may in turn responsible for their different binding individualities. The analysis divulges that both compound 82 and Epirubicin act as potential inhibitors of human telomeric DNA where both ligands stabilizes G4 DNA in presence of K+ ion which was plausibly affect association of telomerase with telomeric DNA therefore exhibiting cell induced apoptosis as an alternate mechanism to damage on binding to quadruplexG4 DNA.
Furthermore, we obtained promising results in this study because compound 82 binds to the FVYL motif/pocket of TERT and adopts the same conformation as the clinical trial agent, BIBR1532. Beside this, acridine ring was established and investigated to be bind with human telomeric G DNA. In light of this rationale, docked analysis of the most active compound 82 in human telomeric G DNA revealed that compound 82 binds to Human telomeric G DNA through hydrogen bonding and hydrophobic interactions. While compound 82 has a different binding conformation than Epirubicin, both exhibit hydrogen bonding and hydrophobic interactions. Based on the docking findings, it is clear that the most active compound 82 binds to both TERT and human telomeric G DNA through similar interactions. As a result, it is reasonable to conclude that compound 82 has potent anticancer activity via dual inhibition of TERT and human telomeric G DNA. The outcome of QSAR and molecular docking study may possibly support many researchers to put forward novel and dual inhibitors of TERT and Human telomeric G DNA with higher anti-cancer activities. Therefore, they may possibly decrease time, cost and even the accessibility of the laboratories equipped to bring out the synthesis and tests.
Availability of data and materials
Data and material are available on request. A dataset of structurally varied 82 nitrogen ring containing heterocycles experimentally tested against Human TERT inhibitory potential has been carefully chosen for QSAR investigation from renown and publicly accessible ChEMBL database (https://www.ebi.ac.uk/chembl/).
Abbreviations
- QSAR:
-
Quantitative structure activity relationship
- GA-MLR:
-
Genetic algorithm multiple linear equation
- TERT:
-
Telomerase reverse transcriptase
- RNP:
-
RNA–protein complex
- MDR:
-
Multidrug resistance
- OECD:
-
Organization for Economic Corporation and Development
- OFS:
-
Objective feature selection
- SFS:
-
Subjective feature selection
- CCC:
-
Concordance correlation coefficient
References
Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33(5):787–791. https://doi.org/10.1016/s0959-8049(97)00062-2
Chen Z, Monia BP, Corey DR (2002) Telomerase inhibition, telomere shortening, and decreased cell proliferation by cell permeable 2′-O-methoxyethyl oligonucleotides. J Med Chem 45(25):5423–5425. https://doi.org/10.1021/jm025563v
Bodnar AG (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279(5349):349–352. https://doi.org/10.1126/science.279.5349.349
Blackburn EH, Collins K (2010) Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harbor Perspect Biol 3(5):a003558-a. https://doi.org/10.1101/cshperspect.a003558
Shay JW (2016) Role of telomeres and telomerase in aging and cancer. Cancer Discov 6(6):584–593. https://doi.org/10.1158/2159-8290.Cd-16-0062
Maida Y, Masutomi K (2015) Telomerase reverse transcriptase moonlights: therapeutic targets beyond telomerase. Cancer Sci 106(11):1486–1492. https://doi.org/10.1111/cas.12806
Ruden M, Puri N (2013) Novel anticancer therapeutics targeting telomerase. Cancer Treatment Rev 39(5):444–456. https://doi.org/10.1016/j.ctrv.2012.06.007
Barma DK, Elayadi A, Falck JR, Corey DR (2003) Inhibition of telomerase by BIBR 1532 and related analogues. Bioorg Med Chem Lett 13(7):1333–1336. https://doi.org/10.1016/s0960-894x(03)00101-x
El-Daly H, Kull M, Zimmermann S, Pantic M, Waller CF, Martens UM (2005) Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood 105(4):1742–1749. https://doi.org/10.1182/blood-2003-12-4322
Yang H, Fu F, Li W, Wei W, Zhang Y, Liu S (2019) Telomerase and poly(ADP-ribose) polymerase-1 activity sensing based on the high fluorescence selectivity and sensitivity of TOTO-1 towards G bases in single-stranded DNA and poly(ADP-ribose). Chem Sci 10(13):3706–3714. https://doi.org/10.1039/c8sc05770b
Saygin C, Carraway HE (2021) Current and emerging strategies for management of myelodysplastic syndromes. Blood Rev 48:100791. https://doi.org/10.1016/j.blre.2020.100791
Hernandez-Meza G, Felden J, Gonzalez-Kozlova EE, Garcia-Lezana T, Peix J, Portela A et al (2021) DNA methylation profiling of human hepatocarcinogenesis. Hepatology 74(1):183–199. https://doi.org/10.1002/hep.31659
Carloni L-E, Wechselberger R, De Vijlder T (2021) Characterization of in vitro G-quadruplex formation of imetelstat telomerase inhibitor. Nucleic Acid Ther. https://doi.org/10.1089/nat.2020.0918
Cheng T, Li Q, Zhou Z, Wang Y, Bryant SH (2012) Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J 14(1):133–141. https://doi.org/10.1208/s12248-012-9322-0
Şekeroğlu ZA, Şekeroğlu V, Küçük N (2020) Effects of reverse transcriptase inhibitors on proliferation, apoptosis, and migration in breast carcinoma cells. Int J Toxicol 40(1):52–61. https://doi.org/10.1177/1091581820961498
Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature 346(6287):866–868. https://doi.org/10.1038/346866a0
Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J 11(5):1921–1929. https://doi.org/10.1002/j.1460-2075.1992.tb05245.x
Kim N, Piatyszek M, Prowse K, Harley C, West M, Ho P (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266(5193):2011–2015. https://doi.org/10.1126/science.7605428
Counter CM, Botelho FM, Wang P, Harley CB, Bacchetti S (1994) Stabilization of short telomeres and telomerase activity accompany immortalization of Epstein–Barr virus-transformed human B lymphocytes. J Virol 68(5):3410–3414. https://doi.org/10.1128/jvi.68.5.3410-3414.1994
Karlseder J (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283(5406):1321–1325. https://doi.org/10.1126/science.283.5406.1321
Karlseder J (2002) Senescence induced by altered telomere state, not telomere loss. Science 295(5564):2446–2449. https://doi.org/10.1126/science.1069523
Blackburn EH, Greider CW, Szostak JW (2006) Telomeres and telomerase: the path from maize, tetrahymena and yeast to human cancer and aging. Nat Med 12(10):1133–1138. https://doi.org/10.1038/nm1006-1133
Cech TR (2004) Beginning to understand the end of the chromosome. Cell 116(2):273–279. https://doi.org/10.1016/s0092-8674(04)00038-8
Han H, Hurley LH (2000) G-quadruplex DNA: a potential target for anti-cancer drug design. Trends Pharmacol Sci 21(4):136–142. https://doi.org/10.1016/s0165-6147(00)01457-7
Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY (2000) G-quadruplexes as targets for drug design. Pharmacol Ther 85(3):141–158. https://doi.org/10.1016/s0163-7258(99)00068-6
Neidle S, Read MA (2000) G-quadruplexes as therapeutic targets. Biopolymers 56(3):195–208. https://doi.org/10.1002/1097-0282(2000)56:3%3c195::Aid-bip10009%3e3.0.Co;2-5
Hurley LH (2001) Secondary DNA structures as molecular targets for cancer therapeutics. Biochem Soc Trans 29(6):692–696. https://doi.org/10.1042/bst0290692
Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T (2006) G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene 25(13):1955–1966. https://doi.org/10.1038/sj.onc.1209217
Shin-Ya K, Wierzba K, Matsuo K-I, Ohtani T, Yamada Y, Furihata K (2001) Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc 123(6):1262–1263. https://doi.org/10.1021/ja005780q
Kim M-Y, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH (2002) Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J Am Chem Soc 124(10):2098–2099. https://doi.org/10.1021/ja017308q
Rosu F, Gabelica V, Shin-Ya K, De Pauw E (2003) Telomestatin-induced stabilization of the human telomeric DNA quadruplex monitored by electrospray mass spectrometry. Chem Commun 21:2702. https://doi.org/10.1039/b309394h
Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T (2003) Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene 22(34):5338–5347. https://doi.org/10.1038/sj.onc.1206833
Gomez D, Paterski R, Lemarteleur T, Shin-Ya K, Mergny J-L, Riou J-F (2004) Interaction of telomestatin with the telomeric single-strand overhang. J Biol Chem 279(40):41487–41494. https://doi.org/10.1074/jbc.M406123200
Perry PJ, Reszka AP, Wood AA, Read MA, Gowan SM, Dosanjh HS (1998) Human telomerase inhibition by regioisomeric disubstituted amidoanthracene-9,10-diones. J Med Chem 41(24):4873–4884. https://doi.org/10.1021/jm981067o
Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO (1997) Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem 40(14):2113–2116. https://doi.org/10.1021/jm970199z
Gowan SM, Heald R, Stevens MFG, Kelland LR (2001) Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol Pharmacol 60(5):981–988. https://doi.org/10.1124/mol.60.5.981
Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S (2002) A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol Pharmacol 61(5):1154–1162. https://doi.org/10.1124/mol.61.5.1154
Harrison RJ, Cuesta J, Chessari G, Read MA, Basra SK, Reszka AP (2003) Trisubstituted acridine derivatives as potent and selective telomerase inhibitors. J Med Chem 46(21):4463–4476. https://doi.org/10.1021/jm0308693
Harrison RJ, Reszka AP, Haider SM, Romagnoli B, Morrell J, Read MA (2004) Evaluation of by disubstituted acridone derivatives as telomerase inhibitors: the importance of G-quadruplex binding. Bioorg Med Chem Lett 14(23):5845–5849. https://doi.org/10.1016/j.bmcl.2004.09.037
Moore MJB, Schultes CM, Cuesta J, Cuenca F, Gunaratnam M, Tanious FA (2006) Trisubstituted acridines as G-quadruplex telomere targeting agents. Effects of extensions of the 3, 6- and 9-side chains on quadruplex binding, telomerase activity, and cell proliferation. J Med Chem 49(2):582–599. https://doi.org/10.1021/jm050555a
Anantha NV, Azam M, Sheardy RD (1998) Porphyrin binding to quadruplexed T4G4. Biochemistry 37(9):2709–2714. https://doi.org/10.1021/bi973009v
Arthanari H, Basu S, Kawano TL, Bolton PH (1998) Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res 26(16):3724–3728. https://doi.org/10.1093/nar/26.16.3724
Haq I, Trent JO, Chowdhry BZ, Jenkins TC (1999) Intercalative G-tetraplex stabilization of telomeric DNA by a cationic porphyrin1. J Am Chem Soc 121(9):1768–1779. https://doi.org/10.1021/ja981554t
Han H, Langley DR, Rangan A, Hurley LH (2001) Selective interactions of cationic porphyrins with G-quadruplex structures. J Am Chem Soc 123(37):8902–8913. https://doi.org/10.1021/ja002179j
Shi D-F, Wheelhouse RT, Sun D, Hurley LH (2001) Quadruplex-interactive agents as telomerase inhibitors: synthesis of porphyrins and structure−activity relationship for the inhibition of telomerase. J Med Chem 44(26):4509–4523. https://doi.org/10.1021/jm010246u
Yamashita T, Uno T, Ishikawa Y (2005) Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg Med Chem 13(7):2423–2430. https://doi.org/10.1016/j.bmc.2005.01.041
Moorhouse AD, Santos AM, Gunaratnam M, Moore M, Neidle S, Moses JE (2006) Stabilization of G-quadruplex DNA by highly selective ligands via click chemistry. J Am Chem Soc 128(50):15972–15973. https://doi.org/10.1021/ja0661919
Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH (1998) NMR-based model of a telomerase-inhibiting compound bound to G-quadruplex DNA†. Biochemistry 37(36):12367–12374. https://doi.org/10.1021/bi981330n
Han H, Cliff CL, Hurley LH (1999) Accelerated assembly of G-quadruplex structures by a small molecule. Biochemistry 38(22):6981–6986. https://doi.org/10.1021/bi9905922
Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM (2006) Evaluation of binding of perylene diimide and benzannulated perylene diimide ligands to DNA by electrospray ionization mass spectrometry. J Am Soc Mass Spectrom 17(4):593–604. https://doi.org/10.1016/j.jasms.2005.12.011
Guo Q, Lu M, Marky LA, Kallenbach NR (2002) Interaction of the dye ethidium bromide with DNA containing guanine repeats. Biochemistry 31(9):2451–2455. https://doi.org/10.1021/bi00124a002
Koeppel F (2001) Ethidium derivatives bind to G-quartets, inhibit telomerase and act as fluorescent probes for quadruplexes. Nucleic Acids Res 29(5):1087–1096. https://doi.org/10.1093/nar/29.5.1087
Perry PJ, Read MA, Davies RT, Gowan SM, Reszka AP, Wood AA (1999) 2,7-Disubstituted amidofluorenone derivatives as inhibitors of human telomerase. J Med Chem 42(14):2679–2684. https://doi.org/10.1021/jm990084q
Mergny JL, Lacroix L, Teulade-Fichou MP, Hounsou C, Guittat L, Hoarau M (2001) Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc Natl Acad Sci 98(6):3062–3067. https://doi.org/10.1073/pnas.051620698
Leonetti C, Amodei S, D’Angelo C, Rizzo A, Benassi B, Antonelli A (2004) Biological activity of the G-quadruplex ligand RHPS4 (3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) is associated with telomere capping alteration. Mol Pharmacol 66(5):1138–1146. https://doi.org/10.1124/mol.104.001537
Cookson JC, Dai F, Smith V, Heald RA, Laughton CA, Stevens MFG (2005) Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate (RHPS4) in vitro: activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol Pharmacol 68(6):1551–1558. https://doi.org/10.1124/mol.105.013300
Skok Ž, Zidar N, Kikelj D, Ilaš J (2019) Dual inhibitors of human DNA topoisomerase II and other cancer-related targets. J Med Chem 63(3):884–904. https://doi.org/10.1021/acs.jmedchem.9b00726
Dallavalle S, Musso L, Artali R, Aviñó A, Scaglioni L, Eritja R (2021) G-Quadruplex binding properties of a potent PARP-1 inhibitor derived from 7-azaindole-1-carboxamide. Sci Rep 11(1):1–13. https://doi.org/10.1038/s41598-021-83474-9
Seimiya H, Nagasawa K, Shin-Ya K (2021) Chemical targeting of G-quadruplexes in telomeres and beyond for molecular cancer therapeutics. J Antibiot. https://doi.org/10.1038/s41429-021-00454-x
Berei J, Eckburg A, Miliavski E, Anderson AD, Miller RJ, Dein J (2020) Potential telomere-related pharmacological targets. Curr Top Med Chem 20(6):458–484. https://doi.org/10.2174/1568026620666200109114339
Singh M, Wang S, Joo H, Ye Z, Christison KM, Hekman R (2020) Use of neomycin as a structured amino-containing side chain motif for phenanthroline-based G-quadruplex ligands and telomerase inhibitors. Chem Biol Drug Des 96(5):1292–1304. https://doi.org/10.1111/cbdd.13741
Xu C-X, Liu L-Y, Lv B, Zhao H-Y, Cao Q, Zhai T (2020) Two novel fan-shaped trinuclear Pt(ii) complexes act as G-quadruplex binders and telomerase inhibitors. Dalton Trans 49(27):9322–9329. https://doi.org/10.1039/d0dt01767a
Chen S, Xue D, Chuai G, Yang Q, Liu Q, Valencia A (2020) FL-QSAR: a federated learning-based QSAR prototype for collaborative drug discovery. Bioinformatics 36(22–23):5492–5498. https://doi.org/10.1093/bioinformatics/btaa1006
Sharma S, Bhatia V (2021) Recent trends in QSAR in modelling of drug–protein and protein–protein interactions. Combin Chem High Throughput Screen 24(7):1031–1041. https://doi.org/10.2174/1386207323666201209093537
Sato A, Miyao T, Jasial S, Funatsu K (2021) Comparing predictive ability of QSAR/QSPR models using 2D and 3D molecular representations. J Comput Aided Mol Des 35(2):179–193. https://doi.org/10.1007/s10822-020-00361-7
Elfadadny A, El-Husseiny HM, Abugomaa A, Ragab RF, Mady EA, Aboubakr M (2021) Role of multidrug resistance-associated proteins in cancer therapeutics: past, present, and future perspectives. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-021-15759-5
Gramatica P (2020) Principles of QSAR modeling. Int J Quant Struct Prop Relationsh 5(3):61–97. https://doi.org/10.4018/IJQSPR.20200701.oa1
Fujita T, Winkler DA (2016) Understanding the roles of the “Two QSARs.” J Chem Inform Model 56(2):269–274. https://doi.org/10.1021/acs.jcim.5b00229
Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M (2014) QSAR modeling: where have you been? Where are you going to? J Med Chem 57(12):4977–5010. https://doi.org/10.1021/jm4004285
Dearden JC, Cronin MTD, Kaiser KLE (2009) How not to develop a quantitative structure–activity or structure–property relationship (QSAR/QSPR). SAR QSAR Environ Res 20(3–4):241–266. https://doi.org/10.1080/10629360902949567
Masand VH, El-Sayed NNE, Mahajan DT, Mercader AG, Alafeefy AM, Shibi IG (2017) QSAR modeling for anti-human African trypanosomiasis activity of substituted 2-phenylimidazopyridines. J Mol Struct 1130:711–718. https://doi.org/10.1016/j.molstruc.2016.11.012
Gramatica P (2014) External evaluation of QSAR models, in addition to cross-validation: verification of predictive capability on totally new chemicals. Mol Inf 33(4):311–314. https://doi.org/10.1002/minf.201400030
Gramatica P (2013) On the development and validation of QSAR models. In: Reisfeld B, Mayeno AN (eds) Computational toxicology. Methods in molecular biology. Humana Press, Totowa, pp 499–526
Huang J, Fan X (2011) Why QSAR fails: an empirical evaluation using conventional computational approach. Mol Pharm 8(2):600–608. https://doi.org/10.1021/mp100423u
Masand VH, El-Sayed NNE, Bambole MU, Patil VR, Thakur SD (2019) Multiple quantitative structure–activity relationships (QSARs) analysis for orally active trypanocidal N-myristoyltransferase inhibitors. J Mol Struct 1175:481–487. https://doi.org/10.1016/j.molstruc.2018.07.080
Masand VH, Rastija V, Patil MK, Gandhi A, Chapolikar A (2020) Extending the identification of structural features responsible for anti-SARS-CoV activity of peptide-type compounds using QSAR modelling. SAR QSAR Environ Res 31(9):643–654. https://doi.org/10.1080/1062936x.2020.1784271
Masand VH, Elsayed NN, Thakur SD, Gawhale N, Rathore MM (2019) Quinoxalinones based aldose reductase inhibitors: 2D and 3D-QSAR analysis. Mol Inform 38(8–9):1800149. https://doi.org/10.1002/minf.201800149
Masand VH, El-Sayed NNE, Bambole MU, Quazi SA (2018) Multiple QSAR models, pharmacophore pattern and molecular docking analysis for anticancer activity of α, β-unsaturated carbonyl-based compounds, oxime and oxime ether analogues. J Mol Struct 1157:89–96. https://doi.org/10.1016/j.molstruc.2017.12.045
Masand VH, El-Sayed NNE, Mahajan DT, Rastija V (2017) QSAR analysis for 6-arylpyrazine-2-carboxamides as Trypanosoma brucei inhibitors. SAR QSAR Environ Res 28(2):165–177. https://doi.org/10.1080/1062936x.2017.1292407
Masand VH, Rastija V (2017) PyDescriptor: a new PyMOL plugin for calculating thousands of easily understandable molecular descriptors. Chemom Intell Lab Syst 169:12–18. https://doi.org/10.1016/j.chemolab.2017.08.003
Masand VH, Mahajan DT, Maldhure AK, Rastija V (2016) Quantitative structure–activity relationships (QSARs) and pharmacophore modeling for human African trypanosomiasis (HAT) activity of pyridyl benzamides and 3-(oxazolo[4,5-b]pyridin-2-yl)anilides. Med Chem Res 25(10):2324–2334. https://doi.org/10.1007/s00044-016-1664-1
Gramatica P, Cassani S, Chirico N (2014) QSARINS-chem: insubria datasets and new QSAR/QSPR models for environmental pollutants in QSARINS. J Comput Chem 35(13):1036–1044. https://doi.org/10.1002/jcc.23576
Gramatica P, Chirico N, Papa E, Cassani S, Kovarich S (2013) QSARINS: a new software for the development, analysis, and validation of QSAR MLR models. J Comput Chem 34(24):2121–2132. https://doi.org/10.1002/jcc.23361
Consonni V, Todeschini R, Ballabio D, Grisoni F (2019) On the misleading use of QF32 for QSAR model comparison. Mol Inform 38(1–2):1800029. https://doi.org/10.1002/minf.201800029
Khanna V, Ranganathan S (2009) Physicochemical property space distribution among human metabolites, drugs and toxins. BMC Bioinform 10(S15):1–18. https://doi.org/10.1186/1471-2105-10-s15-s10
Yuan S, Chan HCS, Hu Z (2017) Using PyMOL as a platform for computational drug design. WIREs Comput Mol Sci 7(2):e1298. https://doi.org/10.1002/wcms.1298
Pennington LD, Moustakas DT (2017) The necessary nitrogen atom: a versatile high-impact design element for multiparameter optimization. J Med Chem 60(9):3552–3579. https://doi.org/10.1021/acs.jmedchem.6b01807
Horvath MP, Schultz SC (2001) DNA G-quartets in a 186 Å resolution structure of an Oxytricha nova telomeric protein-DNA complex. J Mol Biol 310(2):367–377. https://doi.org/10.1006/jmbi.2001.4766
Wang Y, Patel DJ (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1(4):263–282. https://doi.org/10.1016/0969-2126(93)90015-9
Acknowledgements
We are thankful to Shri Yogendraji R. Gode, President, Dr. Rajendra Gode Institute of Pharmacy, University Mardi Road, Amravati, for providing extended support and Research Amenities during entire course of Research work. Authors of this manuscript are thankful to Dr. Paola Gramatica for providing a copy of QSARINS 2.24 software.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization: RDJ and RLB; Formal analysis: CMJ, MSJ and JVM, Data curation and analysis: IL and PNK; Writing and analysis: RDJ. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Authors declares no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Docking results for Compound 82, BABR1532 and Epirubicin into the TERT; Table S2. Docking results for Epirubicin and compound 82 in the human Telomeric G DNA; Table S3. Depiction of QSAR results along with their experimental and predicted EC50 values for divided set model; Table S4. Depiction of QSAR results along with their experimental and predicted EC50 values for full set model; Table S5. Presentation of Serial number, CheMBL id, Smiles and Pec50 value of 82 compounds; Table S6. Calculated Descriptor values used to build QSAR model.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jawarkar, R.D., Bakal, R.L., Khatale, P.N. et al. QSAR, pharmacophore modeling and molecular docking studies to identify structural alerts for some nitrogen heterocycles as dual inhibitor of telomerase reverse transcriptase and human telomeric G-quadruplex DNA. Futur J Pharm Sci 7, 231 (2021). https://doi.org/10.1186/s43094-021-00380-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43094-021-00380-7